Tutorial 8: Masking with One Condition#
Week 1, Day 1, Climate System Overview
Content creators: Sloane Garelick, Julia Kent
Content reviewers: Katrina Dobson, Younkap Nina Duplex, Danika Gupta, Maria Gonzalez, Will Gregory, Nahid Hasan, Paul Heubel, Sherry Mi, Beatriz Cosenza Muralles, Jenna Pearson, Agustina Pesce, Chi Zhang, Ohad Zivan
Content editors: Paul Heubel, Jenna Pearson, Chi Zhang, Ohad Zivan
Production editors: Wesley Banfield, Paul Heubel, Jenna Pearson, Konstantine Tsafatinos, Chi Zhang, Ohad Zivan
Our 2024 Sponsors: CMIP, NFDI4Earth
 #
#
Pythia credit: Rose, B. E. J., Kent, J., Tyle, K., Clyne, J., Banihirwe, A., Camron, D., May, R., Grover, M., Ford, R. R., Paul, K., Morley, J., Eroglu, O., Kailyn, L., & Zacharias, A. (2023). Pythia Foundations (Version v2023.05.01) https://zenodo.org/record/8065851
Tutorial Objectives#
Estimated timing of tutorial: 15 minutes
One useful tool for assessing climate data is masking, which allows you to filter elements of a dataset according to a specific condition and create a “masked array” in which the elements not fulfilling the condition will not be shown. This tool is helpful if you wish to, for example, only look at data greater or less than a certain value, or from a specific temporal or spatial range. For instance, when analyzing a map of global precipitation, we could mask regions that contain a value of mean annual precipitation above or below a specific value or range of values in order to assess wet and dry seasons.
In this tutorial, you will learn how to mask data with one condition and will apply this to your map of global SST.
Setup#
# installations ( uncomment and run this cell ONLY when using google colab or kaggle )
#!pip install pythia_datasets cftime nc-time-axis
# imports
import matplotlib.pyplot as plt
import xarray as xr
from pythia_datasets import DATASETS
Install and import feedback gadget#
Show code cell source
# @title Install and import feedback gadget
!pip3 install vibecheck datatops --quiet
from vibecheck import DatatopsContentReviewContainer
def content_review(notebook_section: str):
return DatatopsContentReviewContainer(
"", # No text prompt
notebook_section,
{
"url": "https://pmyvdlilci.execute-api.us-east-1.amazonaws.com/klab",
"name": "comptools_4clim",
"user_key": "l5jpxuee",
},
).render()
feedback_prefix = "W1D1_T8"
Figure Settings#
Show code cell source
# @title Figure Settings
import ipywidgets as widgets # interactive display
%config InlineBackend.figure_format = 'retina'
plt.style.use(
"https://raw.githubusercontent.com/neuromatch/climate-course-content/main/cma.mplstyle"
)
Video 1: Climate Feedbacks#
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_Climate_Feedbacks_Video")
If you want to download the slides: https://osf.io/download/zhb3c/
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_Climate_Feedbacks_Slides")
Section 1: Masking Data#
Using the xr.where() or .where() method, elements of an Xarray Dataset or Xarray DataArray that satisfy a given condition or multiple conditions can be replaced/masked. To demonstrate this, we are going to use the .where() method on the tos DataArray from the climate model CESM2 that we’ve been using in the past few tutorials.
Let’s load the same data that we used in the previous tutorial (monthly SST data from CESM2):
filepath = DATASETS.fetch("CESM2_sst_data.nc")
ds = xr.open_dataset(filepath)
ds
/opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/xarray/conventions.py:440: SerializationWarning: variable 'tos' has multiple fill values {1e+20, 1e+20}, decoding all values to NaN.
new_vars[k] = decode_cf_variable(
<xarray.Dataset> Size: 47MB
Dimensions: (time: 180, d2: 2, lat: 180, lon: 360)
Coordinates:
* time (time) object 1kB 2000-01-15 12:00:00 ... 2014-12-15 12:00:00
* lat (lat) float64 1kB -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 3kB 0.5 1.5 2.5 3.5 4.5 ... 356.5 357.5 358.5 359.5
Dimensions without coordinates: d2
Data variables:
time_bnds (time, d2) object 3kB ...
lat_bnds (lat, d2) float64 3kB ...
lon_bnds (lon, d2) float64 6kB ...
tos (time, lat, lon) float32 47MB ...
Attributes: (12/45)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 972
... ...
sub_experiment_id: none
table_id: Omon
tracking_id: hdl:21.14100/2975ffd3-1d7b-47e3-961a-33f212ea4eb2
variable_id: tos
variant_info: CMIP6 20th century experiments (1850-2014) with C...
variant_label: r11i1p1f1Section 1.1: Using .where() with One Condition#
Let’s say we want to analyze SST just from the last time in the dataset (2014-09-15). We can isolate this time using .isel():
sample = ds.tos.sel(time='2014-09')
sample
<xarray.DataArray 'tos' (time: 1, lat: 180, lon: 360)> Size: 259kB
[64800 values with dtype=float32]
Coordinates:
* time (time) object 8B 2014-09-15 00:00:00
* lat (lat) float64 1kB -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 3kB 0.5 1.5 2.5 3.5 4.5 ... 356.5 357.5 358.5 359.5
Attributes: (12/19)
cell_measures: area: areacello
cell_methods: area: mean where sea time: mean
comment: Model data on the 1x1 grid includes values in all cells f...
description: This may differ from "surface temperature" in regions of ...
frequency: mon
id: tos
... ...
time_label: time-mean
time_title: Temporal mean
title: Sea Surface Temperature
type: real
units: degC
variable_id: tosNow that we have our DataArray from the desired time period, we can use another function, .where() documentation to filter elements according to a condition. The conditional expression in .where() can be a DataArray, a Dataset, or a function. Indexing methods on Xarray objects generally return a subset of the original data. However, it is sometimes useful to select an object with the same shape as the original data, but with some elements masked. Unlike .isel() and .sel() which change the shape of the returned results, .where() preserves the shape of the original data. It accomplishes this by returning values from the original DataArray or Dataset if the condition is True, and fills in values (by default nan) wherever the condition is False. Additional information can be found in the .where() documentation.
Let’s use .where() to mask locations with temperature values greater than 0ºC. Note that the condition we supply to .where() is the regions we wish to preserve, not those we wish to mask. So if we are interested in masking temperature values that are above 0ºC, we will pass the condition to preserve those that are less than or equal to 0ºC.
# mask temperatures greater than or equal to 0, preserve those that are less than 0
masked_sample = sample.where(sample <= 0.0)
masked_sample
<xarray.DataArray 'tos' (time: 1, lat: 180, lon: 360)> Size: 259kB
array([[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[-1.6132288, -1.6125063, -1.611589 , ..., -1.6145128,
-1.6141783, -1.6138089],
[-1.597154 , -1.5969657, -1.5967476, ..., -1.5973103,
-1.5972834, -1.5972393],
[-1.5958687, -1.5960269, -1.5961952, ..., -1.5955368,
-1.5956477, -1.5957516]]], dtype=float32)
Coordinates:
* time (time) object 8B 2014-09-15 00:00:00
* lat (lat) float64 1kB -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 3kB 0.5 1.5 2.5 3.5 4.5 ... 356.5 357.5 358.5 359.5
Attributes: (12/19)
cell_measures: area: areacello
cell_methods: area: mean where sea time: mean
comment: Model data on the 1x1 grid includes values in all cells f...
description: This may differ from "surface temperature" in regions of ...
frequency: mon
id: tos
... ...
time_label: time-mean
time_title: Temporal mean
title: Sea Surface Temperature
type: real
units: degC
variable_id: tosLet’s plot both our original sample, and the masked sample for September 15th, 2014. Note we are using a different colorbar for the right hand figure, where the range of values is much smaller, and the same colors on the left would not correspond to the same colors on the right.
fig, axes = plt.subplots(ncols=2, figsize=(19, 6))
sample.plot(ax=axes[0])
masked_sample.plot(ax=axes[1])
<matplotlib.collections.QuadMesh at 0x7ff94912be50>
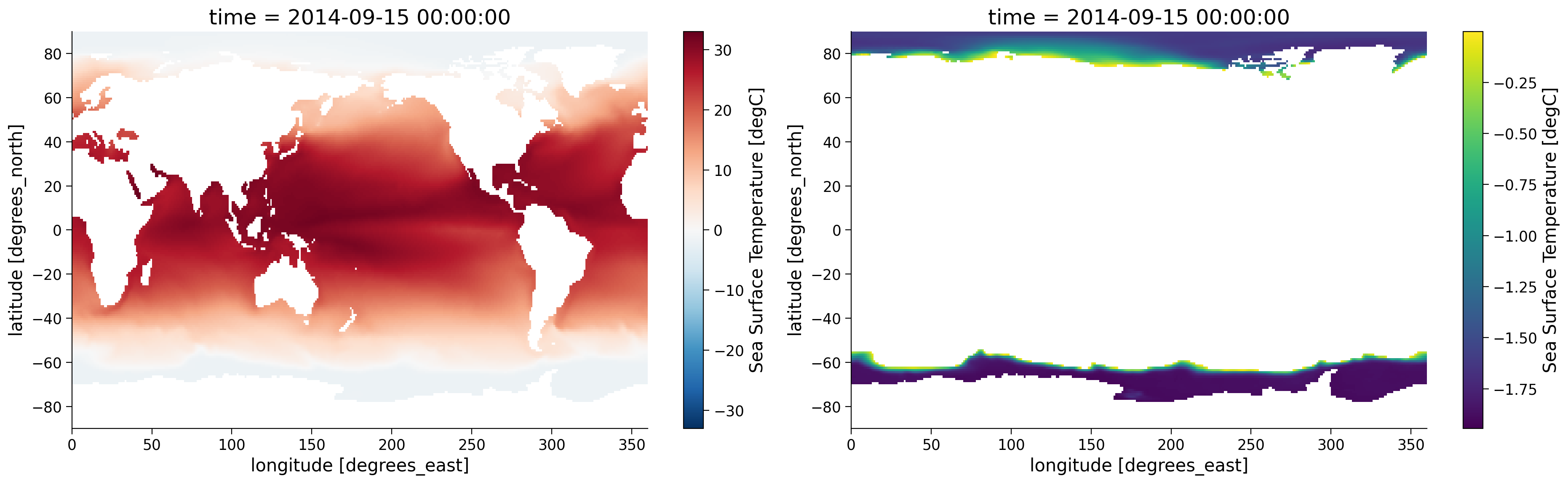
Notice how in the figure on the right, only the SST from the areas where SST is below 0ºC is shown and the other areas are white since these are now NaN values. Now let’s assess how polar SST has changed over the time period recorded by the original dataset. To do so, we can run the same code but focus on the time 2000-09-15.
# create a second sample of September 2000
sample_2 = ds.tos.sel(time="2000-09")
# mask it
masked_sample_2 = sample_2.where(sample_2 < 0.0)
# plot both samples next to each other
fig, axes = plt.subplots(ncols=2, figsize=(19, 6))
masked_sample_2.plot(ax=axes[0])
masked_sample.plot(ax=axes[1])
<matplotlib.collections.QuadMesh at 0x7ff949048190>
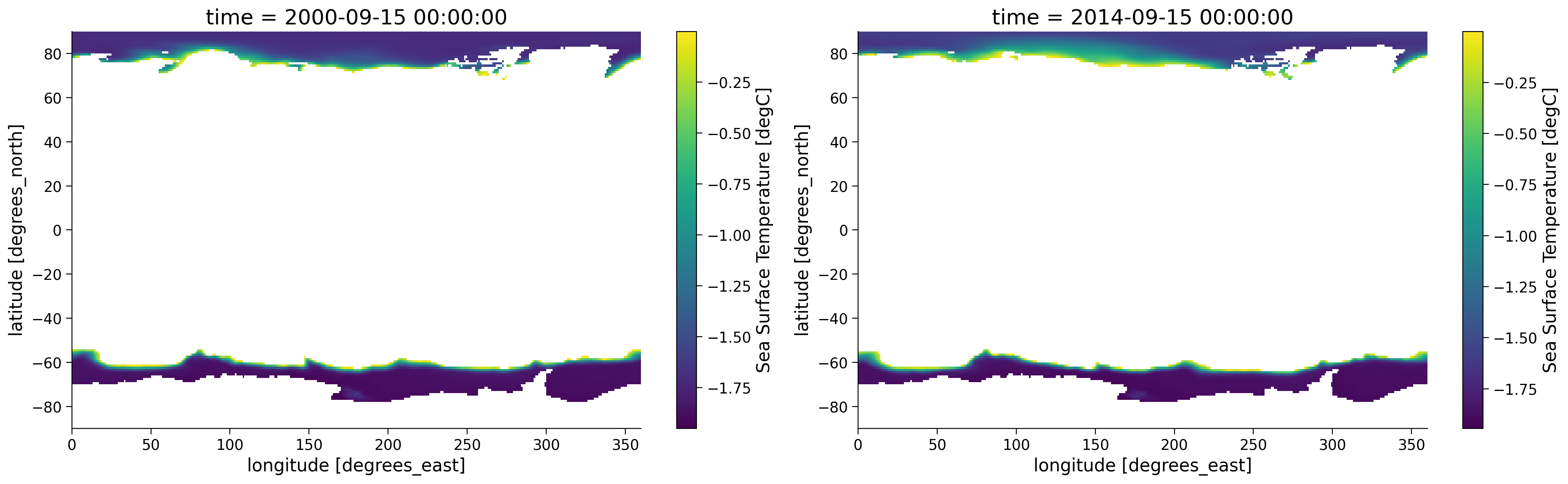
Questions 1.1: Climate Connection#
What is the purpose of masking in the analysis of climate data?
Within the areas that are not masked, how does the distribution of SST compare between these maps?
The minimum sea ice extent in the Arctic typically occurs annually in September after spring has brought in more sunlight and warmer temperatures. Considering both plots of September SST above (from 2000 on the left and 2014 on the right), how might changes in the ice-albedo feedback be playing a role in what you observe? Please state any assumptions you would make in your answer.
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_Questions_1_1")
Summary#
In this tutorial, we’ve explored the application of masking tools in the analysis of Sea Surface Temperature (SST) maps. Through masking, we’ve been able to focus our attention on areas where the SST is below 0°C. These are the regions where changes in the ice-albedo feedback mechanism are most evident in our present day. This has facilitated a more targeted analysis and clearer understanding of the data.
Resources#
Code and data for this tutorial is based on existing content from Project Pythia.



